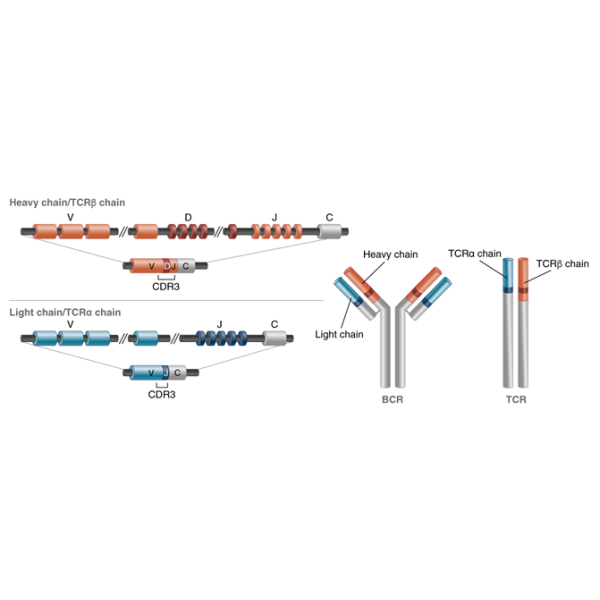
The NEBNext Immune Sequencing Kit (Mouse) enables exhaustive profiling of somatic mutations in the full-length immune gene repertoires of B cells and T cells, via the expression of complete antibody chains. This includes modular primer sets, providing information for complete V, D, and J segments and full isotype information analysis (IgM, IgD, IgG, IgA and IgE), and TCRα, TCRβ, TCRγ and TCRδ chain characterization. A unique, UMI-based mRNA barcoding process allows PCR copies derived from an individual molecule to be converted to a consensus sequence. This improves sequence accuracy and eliminates PCR bias. Based on the open-source pRESTO toolkit, a workflow is available via the Galaxy platform to enable robust bioinformatic analysis locally or in a cloud environment. A workflow tutorial ensures that users who are not familiar with Galaxy can successfully use the analysis workflow,
Immune repertoire sequencing is frequently used to analyze immune responses, both current and distant. Areas of particular interest include characterization of autoimmune diseases, oncology, discovery of neutralizing antibodies against infectious disease, tumor-infiltrating lymphocytes and use as a tool to study residual disease. Recent improvements in read lengths and throughputs of next-generation sequencing (NGS) platforms have resulted in a rise in the popularity of immune repertoire sequencing.
Defining features of the NEBNext Immune Sequencing method include:
- Generation of full-length variable sequences (including isotype information), allowing downstream antibody synthesis and functional characterization not possible with approaches sequencing only the CDR3 region
- Eliminated use of variable region primers, reducing primer pool complexity and realizing unbiased and simultaneous recovery of B cell and T cell receptor transcripts.
- Minimized PCR bias and improved sequencing accuracy by allowing a consensus to be generated from duplicate sequencing reads originating from the same transcript; UMIs enable accurate quantitation of each clone present in the sample.
- Optimized high target-capture efficiency for immune repertoire sequencing and analysis from sub-microgram quantities of total RNA.
Mai multe informatii
| Price | 38.616,00 RON (preturile sunt fara TVA) |
|---|---|
| Description |
The NEBNext Immune Sequencing Kit (Mouse) enables exhaustive profiling of somatic mutations in the full-length immune gene repertoires of B cells and T cells, via the expression of complete antibody chains. This includes modular primer sets, providing information for complete V, D, and J segments and full isotype information analysis (IgM, IgD, IgG, IgA and IgE), and TCRα, TCRβ, TCRγ and TCRδ chain characterization. A unique, UMI-based mRNA barcoding process allows PCR copies derived from an individual molecule to be converted to a consensus sequence. This improves sequence accuracy and eliminates PCR bias. Based on the open-source pRESTO toolkit, a workflow is available via the Galaxy platform to enable robust bioinformatic analysis locally or in a cloud environment. A workflow tutorial ensures that users who are not familiar with Galaxy can successfully use the analysis workflow,
|

 English
English