M0348L, NEBNext dsDNA Fragmentase - 250 assays
NEBNext dsDNA Fragmentase is an enzyme-based reagent that shears DNA to produce fragments of the desired sizes in a time-dependent manner, for next generation sequencing library preparation protocols.
NEBNext dsDNA Fragmentase is an enzyme-based reagent that shears DNA to produce fragments of the desired sizes in a time-dependent manner, for next generation sequencing library preparation protocols.
- dsDNA Fragmentase provides random fragmentation, similar to mechanical methods
Note: NEBNext dsDNA Fragmentase is not part of the NEBNext Ultra™ II FS (Fragmentation System) reagents.
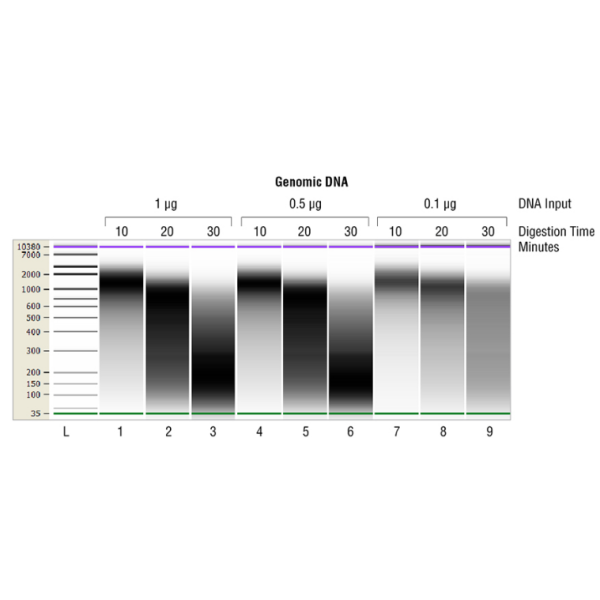
NEBNext® dsDNA Fragmentase generates dsDNA breaks in a time-dependent manner to yield 50–1,000 bp DNA fragments depending on reaction time. NEBNext dsDNA Fragmentase contains two enzymes, one randomly generates nicks on dsDNA and the other recognizes the nicked site and cuts the opposite DNA strand across from the nick, thereby producing dsDNA breaks. The resulting DNA fragments contain short overhangs, 5´-phosphates, and 3´-hydroxyl groups. The random nicking activity of NEBNext dsDNA Fragmentase has been confirmed by preparation of libraries for next-generation sequencing. A comparison of the sequencing results between libraries prepared with genomic DNA sheared with NEBNext dsDNA Fragmentase and with mechanical shearing demonstrates that NEBNext dsDNA Fragmentase does not introduce any detectable bias during sequencing library preparation and no difference in sequence coverage is observed between the two methods.
| Price | 2.970,00 RON (preturile sunt fara TVA) |
|---|---|
| Description |
NEBNext dsDNA Fragmentase is an enzyme-based reagent that shears DNA to produce fragments of the desired sizes in a time-dependent manner, for next generation sequencing library preparation protocols.
Note: NEBNext dsDNA Fragmentase is not part of the NEBNext Ultra™ II FS (Fragmentation System) reagents. NEBNext® dsDNA Fragmentase generates dsDNA breaks in a time-dependent manner to yield 50–1,000 bp DNA fragments depending on reaction time. NEBNext dsDNA Fragmentase contains two enzymes, one randomly generates nicks on dsDNA and the other recognizes the nicked site and cuts the opposite DNA strand across from the nick, thereby producing dsDNA breaks. The resulting DNA fragments contain short overhangs, 5´-phosphates, and 3´-hydroxyl groups. The random nicking activity of NEBNext dsDNA Fragmentase has been confirmed by preparation of libraries for next-generation sequencing. A comparison of the sequencing results between libraries prepared with genomic DNA sheared with NEBNext dsDNA Fragmentase and with mechanical shearing demonstrates that NEBNext dsDNA Fragmentase does not introduce any detectable bias during sequencing library preparation and no difference in sequence coverage is observed between the two methods. |

 English
English