E3350L, NEBNext Enzymatic 5hmC-seq Kit - 96 assays
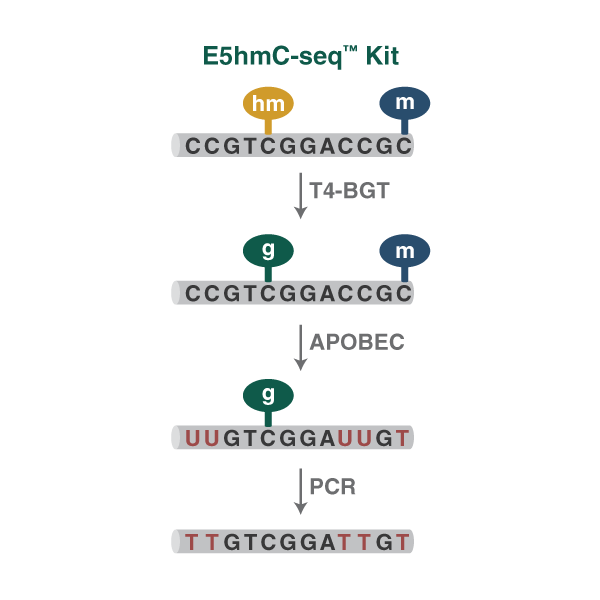
NEBNext Enzymatic 5hmC-seq (E5hmC-seq) uses an enzyme-based method to specifically detect 5hmC, without the DNA damage typical of bisulfite-based methods, and this sensitive method can be used with as little as 0.1 ng of input DNA.
Typically, the modified cytosines 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC) are detected by sequencing of Illumina libraries generated using the NEBNext EM-seq enzyme-based workflow or bisulfite conversion. However, these methods cannot differentiate between 5mC and 5hmC.
As with EM-seq, NEBNext Enzymatic 5hmC-seq (E5hmC-seq) uses an enzyme-based method to specifically detect 5hmC, without the DNA damage typical of bisulfite-based methods, and this sensitive method can be used with as little as 0.1 ng of input DNA.
In a two-step process, T4-BGT glucosylates 5hmC, providing protection from deamination by APOBEC in the next step. T4-BGT does not protect 5mC or unmethylated cytosines, which are deaminated by APOBEC to uracils. This is followed by amplification using a NEBNext master mix formulation of Q5U® (a modified version of Q5® High-Fidelity DNA Polymerase), and sequencing on the Illumina platform.
Bioinformatic analysis tools used for EM-seq and for bisulfite sequencing can also be used for E5hmC-seq. E5hmC-seq data can be subtracted from EM-seq data, thereby allowing determination of the precise location of individual 5mC and 5hmC sites.
Note that index primers are not included and must be purchased separately (NEBNext Primers for Epigenetics, NEB #E3392, NEB #E3404).
Mai multe informatii
| Price | 18.660,00 RON (preturile sunt fara TVA) |
|---|---|
| Description |
Typically, the modified cytosines 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC) are detected by sequencing of Illumina libraries generated using the NEBNext EM-seq enzyme-based workflow or bisulfite conversion. However, these methods cannot differentiate between 5mC and 5hmC. |

 English
English