E8210S, Ph.D.-12 Phage Display Peptide Library Kit v2 - 10 Panning Experiments
The Ph.D.-12 Phage Display Peptide Library Kit v2 is based on a combinatorial library of random dodecapeptides fused to the N-terminus of a minor coat protein (pIII) of M13 phage. The peptide is followed by a short spacer (Gly-Gly-Gly) which directly precedes the wild-type pIII sequence. The library consists of ~109 electroporated sequences amplified once to yield approximately 100 copies of each sequence in 10 μl of the supplied phage.
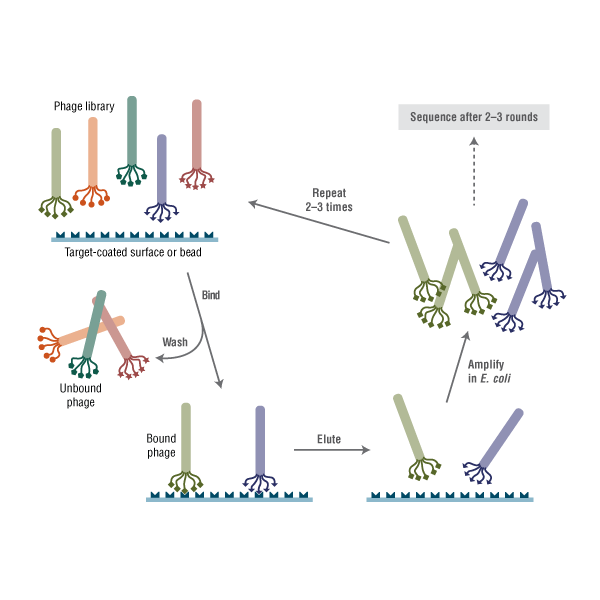
Phage display describes a selection technique in which a library of peptide or protein variants is expressed on the outside of a phage virion, while the genetic material encoding each variant resides on the inside. This creates a physical linkage between each variant protein sequence and the DNA encoding it, which allows rapid partitioning based on binding affinity to a given target molecule (antibodies, enzymes, cell-surface receptors, etc.) by an in vitro selection process called panning. In its simplest form, panning is carried out by incubating a library of phage-displayed peptides on a plate (or bead) coated with the target, washing away the unbound phage, and eluting the specifically bound phage. The eluted phage are then amplified and taken through additional binding/amplification cycles to enrich the pool in favor of binding sequences. After 3–4 rounds, individual clones are characterized by DNA sequencing and binding assays.
The Ph.D.-12 Phage Display Peptide Library Kit v2 is based on a combinatorial library of random dodecapeptides fused to the N-terminus of a minor coat protein (pIII) of M13 phage. The peptide is followed by a short spacer (Gly-Gly-Gly) which directly precedes the wild-type pIII sequence. The library consists of ~109 electroporated sequences amplified once to yield approximately 100 copies of each sequence in 10 μl of the supplied phage.
Features
- Ready to use complex phage library; inherent link between phenotype and genotype allows screening of billions of clones in a single microtiter well or Eppendorf tube
- Control panning experiment does not require blocking step and eliminates spurious plastic binders
- Control panning experiment with Protein G Magnetic Beads reduces tedium of plate/well washing steps
- Length of random region (12-mer) allows for intramolecular secondary structure formation as well as sequence gaps between defined positions
- Does not require helper phage for amplification
- Discover peptide binders in less than one week with straight-forward molecular biology and sterile culture techniques
- Supplied -96 gIII Sequencing Primer (500 pmol) allows for >50 sequencing reactions
Mai multe informatii
| Price | 4.680,00 RON (preturile sunt fara TVA) |
|---|---|
| Description |
Phage display describes a selection technique in which a library of peptide or protein variants is expressed on the outside of a phage virion, while the genetic material encoding each variant resides on the inside. This creates a physical linkage between each variant protein sequence and the DNA encoding it, which allows rapid partitioning based on binding affinity to a given target molecule (antibodies, enzymes, cell-surface receptors, etc.) by an in vitro selection process called panning. In its simplest form, panning is carried out by incubating a library of phage-displayed peptides on a plate (or bead) coated with the target, washing away the unbound phage, and eluting the specifically bound phage. The eluted phage are then amplified and taken through additional binding/amplification cycles to enrich the pool in favor of binding sequences. After 3–4 rounds, individual clones are characterized by DNA sequencing and binding assays. Features
|

 English
English